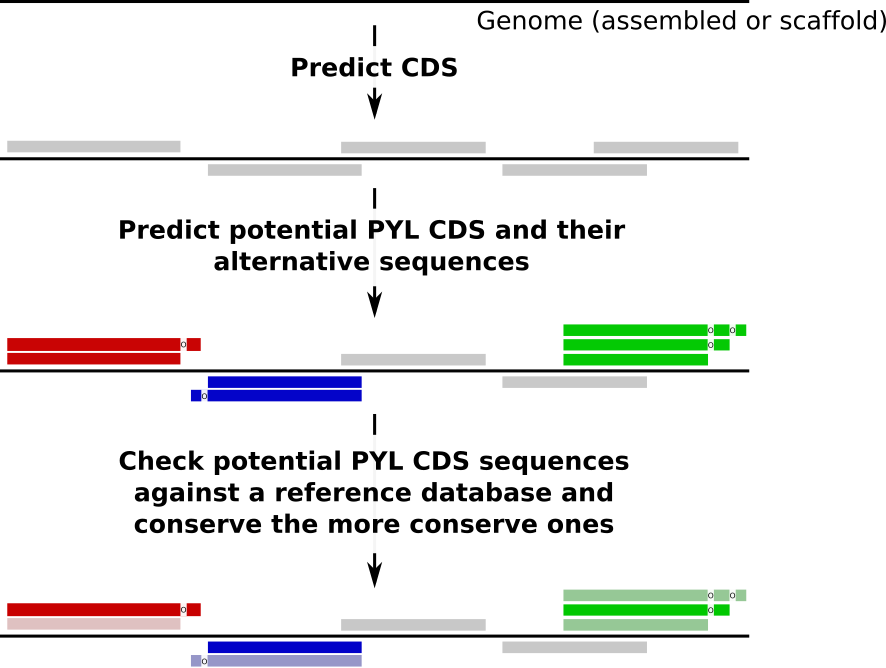
Identification of potential PYL proteins¶
PylProtPredictor identifies proteins potentially using Pyrrolysine as amino acid in any genome, given the following workflow:
Prediction¶
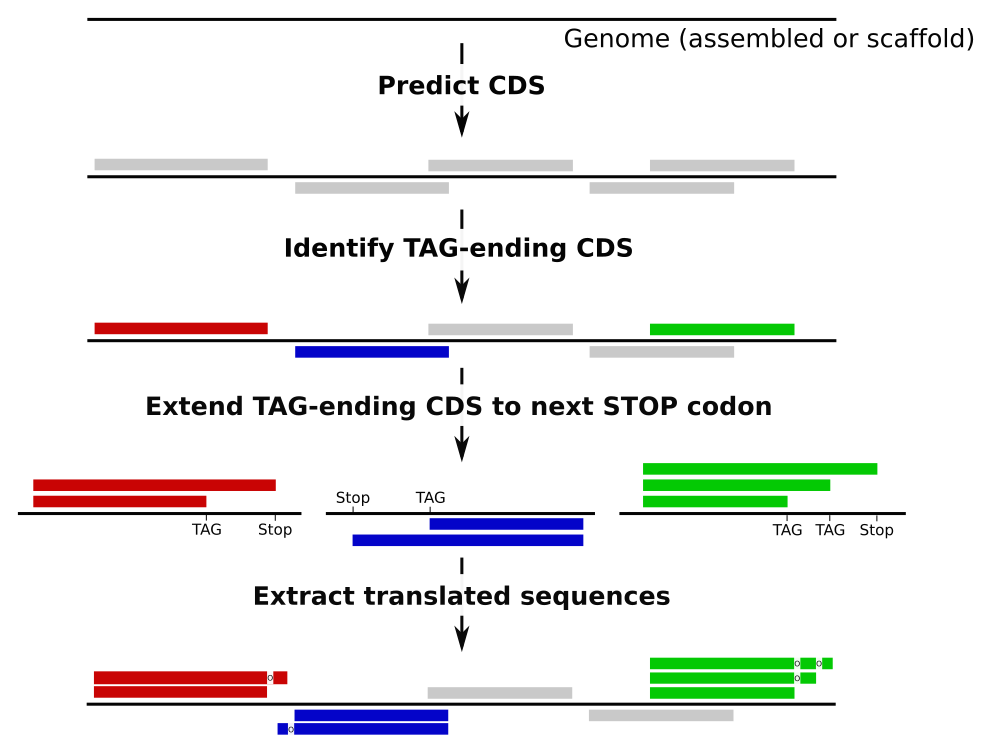
The first step of the workflow is the prediction of proteins potentially using Pyrrolysine (PYL) as amino acid. In these proteins, the TAG codon is not used as a STOP codon but translated into PYL amino acid.
This prediction starts with the prediction of the CDS on a genome using Prodigal. The TAG-ending CDS are then identified: their sequence is extracted and potentially extended until next STOP codon to extract potential alternative sequences for these proteins with PYL as amino acid.
Checking¶
For each TAG-ending CDS, several potential sequences are extracted: the original identified CDS and possible extensions in which TAG would not be a STOP codon but a PYL.
To identify which sequence for each CDS is the most likely, the sequences are searched against a reference database (UniRef 90 by default) using diamond. To identify which sequence for each CDS to converse or reject, the diamond report is parsed. For each potential PYL CDS, an extension is conserved if the evalue of its best match is smaller than the evalue of the best match of the original sequences and if the alignment is longer. It prevents cases where the alignment was similar but the evalue smaller for the extension.